Introduction
MerKurio was developed as command line tool and provides two main functionalities through dedicated subcommands.
The extract subcommand identifies and extracts sequence records from fasta or fastq files based on query k-mers, with full support for paired-end reads where matching in one read extracts both mates.
The tag subcommand annotates sequences in SAM or BAM alignment formats according to the k-mers that they contain following the SAM Optional Fields Specification with user-defined labels (default “km”).
It was developed to sinmplify downstream analysis of selected k-mers by tracing them back to their sequences in the original data.
For installation instructions, go to the installation section.
To test MerKurio on a minimal working example, visit the minimal example section.
For an example workflow, visit the practical tutorial section.
You can use the search bar on top to find specific information, or use the navigation on the left to browse the manual.
If you have found a bug or have a feature request, please open an issue on the GitHub repository or check the discussions on GitHub.
License
The code in this repository is licensed under the MIT license.
Test data and example files in the tests/ and example-minimal/ directories are licensed under the CC0 1.0 Universal license.
Installation
You can install MerKurio in several ways, depending on your system and whether you have Rust installed.
1. Precompiled Binaries (No Rust Needed)
2. Install via Cargo (Requires Rust)
3. Build Manually Without Installing (Requires Rust)
After installation, verify if it works by running:
merkurio --help
Or, if you didn’t add it to your PATH:
./path/to/merkurio --help
Option 1: Precompiled Binaries (No Rust Needed)
Download a binary for Linux, Windows, or macOS from the releases page, then extract the archive:
tar -xzf path/to/release.tar.gz
On Linux/macOS, make it executable if needed:
chmod u+x path/to/merkurio
The merkurio-x86_64-unknown-linux-musl is compatible with a wider range of systems but can have worse performance.
Option 2: Install via Cargo (Requires Rust)
If you have Rust installed (edition 2024), the easiest way is:
cargo install merkurio
This pulls the latest version from crates.io.
To install a tagged release from GitHub:
cargo install --git https://github.com/lschoenm/MerKurio --tag vX.X.X
Option 3: Build Manually Without Installing (Requires Rust)
git clone https://github.com/lschoenm/MerKurio
cd MerKurio
cargo build --release
The binary will be in target/release/.
Minimal Example
Download the files kmers.txt, sample.sam, and sample.fasta from the repository.
After sucessfully installing MerKurio (follow the documentation) and having it added to your PATH, run the following commands to test MerKurio:
# Run MerKurio extract
echo "Extracting sequences in sample.fasta containing the k-mer in kmers.txt:"
merkurio extract -f kmers.txt -i sample.fasta
# Run MerKurio tag
echo "Tagging records in sample.sam with the k-mer in kmers.txt:"
merkurio tag -f kmers.txt -i sample.sam
# Generate only a plain text log
echo "Generating a plain text log of matching statistics:"
merkurio extract -f kmers.txt -i sample.fasta -S -l
# Generate only a JSON log
echo "Generating a JSON log of matching statistics:"
merkurio tag -f kmers.txt -i sample.sam -S -j
Explanation of the input files:
kmers.txt: contains a single k-mer sequence.sample.sam: a synthetic example SAM file, containing five records.sample.fasta: a synthetic FASTA file, containing two sequences.
In this minimal example, no output files are generated. The records and matching statistics are directly written to the terminal. For a description of the plain text log and JSON log, go to the official documentation.
For a detailed explanation of the available parameters of the tag and extract subcommands visit their sections in the documentation.
Example Workflow
A practical example/tutorial can be found in the repository or on the next page.
Although the tool is designed to be flexible and can be used in a variety of ways, it was designed with the following workflow in mind:
- Have a list of k-mers of interest.
- Use MerKurio to extract the FASTA/FASTQ records containing these k-mers (and their reverse complements) from the original (paired-end) records.
If sequencing reads were extracted:
- Align the extracted reads to a reference genome.
- Use MerKurio to tag the aligned records in a SAM/BAM file with the significant k-mers they contain (and optionally filtering it).
- Analyzing the matching statistics generated by MerKurio.
- Interpret the results: e. g., by visualizing the k-mers that are frequently tagged in the reads that align to a particular region of the genome.
Practical example/tutorial for MerKurio
This is a brief demonstration and guide on how MerKurio can be used in practice.
Note: this tutorial is written for Unix-like systems.
The guide is also available in the repository, along with the input and output data.
This tutorial assumes that you have already installed MerKurio on your system. If you haven’t done this yet, please refer to the installation section in the documentation. Additionally, the following software is used in this example:
- jq (v1.6)
- Bowtie2 (v2.5.4)
- samtools (v1.22)
- IGV (v2.19.4)
These dependencies are only necessary for parts of the tutorial after read extraction.
In this guide, we will walk through the following scenario: we have a list of relevant k-mers which we have obtained from a fictional association test. We then want to extract all raw sequencing reads from a mutant strain which contain our k-mers of interest, using MerKurio. Next, we want to map them to the wildtype reference genome. With MerKurio, we can then tag the aligned reads with the k-mers they contain and visualize this information in a genome browser.
Setup
To install MerKurio, follow the installation instructions in the Readme or the official documentation.
If you have cargo/Rust installed, you can first clone the repository using git. This will download a local copy of the repository to your computer. Then you can install MerKurio from source, which will automatically add it to your PATH:
git clone https://github.com/lschoenm/MerKurio
cd MerKurio
cargo install --path .
Test the installation by running MerKurio:
merkurio --help
Alternatively, download a standalone binary from the releases page. If you don’t add merkurio to your PATH, you have to call it using the path to the executable.
Download the data
A full dataset of Staphylococcus aureus is available from Zenodo/the Galaxy training network under this link. It comprises the reference genome sequence in FASTA format, a list of feature annotations in GFF format, and paired-end sequence reads in FASTQ format.
First, let’s create a new directory data and download the files by entering the following commands in your terminal:
mkdir data
curl https://zenodo.org/records/582600/files/mutant_R1.fastq > data/mutant_R1.fastq
curl https://zenodo.org/records/582600/files/mutant_R2.fastq > data/mutant_R2.fastq
curl https://zenodo.org/records/582600/files/wildtype.fna > data/wildtype.fna
curl https://zenodo.org/records/582600/files/wildtype.gff > data/wildtype.gff
The dataset contains the following files:
mutant_R[12].fastqare the two FASTQ files containing the paired-end sequencing reads (~150 bp long).wildtype.fnacontains an assembled miniature genome of S. aureus.wildtype.gfflisting the features of the reference genome.
The genome is from an imaginary S. aureus bacterium. The sequencing reads have around 19x coverage.
Extract sequencing reads based on k-mers
Let’s assume that we ran a k-mer-based genome-wide association study in S. aureus which identified the following k-mers to be significantly associated with antibiotics resistance:
AATATAATAAATTAACCGAAGATAAAAAAGAATCTCAGCACATAAATAATGTGGCAGCACAAATTGTGCTGCCACATTATTTATGTGCTGAGA
Normally, we would assume that we already have those sequences obtained from previous analyses either in a text file or a FASTA file - both of which are valid input for MerKurio. For this example, we have to createa file significant_kmers.txt storing these k-mers, one per line. This can be done with the following commands:
echo AATATAATAAATTAACCGAAGATAAAAAAGA > data/significant_kmers.txt
echo ATCTCAGCACATAAATAATGTGGCAGCACAA >> data/significant_kmers.txt
echo ATTGTGCTGCCACATTATTTATGTGCTGAGA >> data/significant_kmers.txt
We can use MerKurio’s extract subcommand to extract all raw sequencing reads from the mutant S. aureus strain which contain any of these k-mers.
The -o flag controls the output file suffix, -i the path to the first input file. We want to extract read pairs if any of the mates contains one of our k-mers. MerKurio automatically switches to paired-end read mode if the second file is provided with the -2 flag.
Because we do not know from which of the two DNA strands the k-mers are coming yet both of them could be represented in the reads, we also have to look for the reverse complements of the query sequences (this is controlled by the -r flag).
Additionally, we would also like to generate detailed match statistics describing which k-mers were found and their exact locations in the reads, along with a few summary statistics. By providing the -l flag without an argument, a log in plain text format is written to the stdout channel of the terminal. Additionally, we can store a log in JSON format by using the -j flag with a file path.
mkdir output
mkdir logs
merkurio extract -i data/mutant_R1.fastq -2 data/mutant_R2.fastq -f data/significant_kmers.txt -r -o output/mutant_extracted -l -j logs/mutant_extracted.stats.json
Here, you can see the statistics log output:
#MerKurio extract log
#2025-06-27T14:54:18+02:00[Europe/Vienna]
#Running merkurio version 1.0.0
#Command line: merkurio extract -i mutant_R1.fastq -2 mutant_R2.fastq -f significant_kmers.txt -r -o mutant_extracted -l -j logs/mutant_extracted.stats.json
#Searching for 6 patterns
#
#File Record Pattern Position (zero-based)
mutant_R2.fastq mutant-no_snps.gff-23516/2 ATCTCAGCACATAAATAATGTGGCAGCACAA 30
mutant_R2.fastq mutant-no_snps.gff-23516/2 TCTCAGCACATAAATAATGTGGCAGCACAAT 31
mutant_R2.fastq mutant-no_snps.gff-21992/2 ATTGTGCTGCCACATTATTTATGTGCTGAGA 52
mutant_R2.fastq mutant-no_snps.gff-21992/2 TTGTGCTGCCACATTATTTATGTGCTGAGAT 53
mutant_R2.fastq mutant-no_snps.gff-21086/2 ATCTCAGCACATAAATAATGTGGCAGCACAA 113
mutant_R2.fastq mutant-no_snps.gff-21086/2 TCTCAGCACATAAATAATGTGGCAGCACAAT 114
mutant_R2.fastq mutant-no_snps.gff-21076/2 TCTTTTTTATCTTCGGTTAATTTATTATATT 88
mutant_R2.fastq mutant-no_snps.gff-20088/2 ATCTCAGCACATAAATAATGTGGCAGCACAA 108
mutant_R2.fastq mutant-no_snps.gff-20088/2 TCTCAGCACATAAATAATGTGGCAGCACAAT 109
mutant_R2.fastq mutant-no_snps.gff-18432/2 AATATAATAAATTAACCGAAGATAAAAAAGA 35
mutant_R2.fastq mutant-no_snps.gff-15850/2 TCTTTTTTATCTTCGGTTAATTTATTATATT 29
mutant_R2.fastq mutant-no_snps.gff-14898/2 AATATAATAAATTAACCGAAGATAAAAAAGA 75
mutant_R1.fastq mutant-no_snps.gff-14434/1 TCTTTTTTATCTTCGGTTAATTTATTATATT 66
mutant_R2.fastq mutant-no_snps.gff-13984/2 ATTGTGCTGCCACATTATTTATGTGCTGAGA 113
mutant_R2.fastq mutant-no_snps.gff-13984/2 TTGTGCTGCCACATTATTTATGTGCTGAGAT 114
mutant_R1.fastq mutant-no_snps.gff-12520/1 ATCTCAGCACATAAATAATGTGGCAGCACAA 0
mutant_R1.fastq mutant-no_snps.gff-12520/1 TCTCAGCACATAAATAATGTGGCAGCACAAT 1
mutant_R1.fastq mutant-no_snps.gff-10584/1 AATATAATAAATTAACCGAAGATAAAAAAGA 3
mutant_R1.fastq mutant-no_snps.gff-10384/1 TCTTTTTTATCTTCGGTTAATTTATTATATT 25
mutant_R2.fastq mutant-no_snps.gff-8582/2 ATTGTGCTGCCACATTATTTATGTGCTGAGA 32
mutant_R2.fastq mutant-no_snps.gff-8582/2 TTGTGCTGCCACATTATTTATGTGCTGAGAT 33
mutant_R2.fastq mutant-no_snps.gff-8390/2 TCTTTTTTATCTTCGGTTAATTTATTATATT 14
mutant_R2.fastq mutant-no_snps.gff-7378/2 TCTTTTTTATCTTCGGTTAATTTATTATATT 14
mutant_R2.fastq mutant-no_snps.gff-6398/2 ATTGTGCTGCCACATTATTTATGTGCTGAGA 76
mutant_R2.fastq mutant-no_snps.gff-6398/2 TTGTGCTGCCACATTATTTATGTGCTGAGAT 77
mutant_R1.fastq mutant-no_snps.gff-5520/1 ATCTCAGCACATAAATAATGTGGCAGCACAA 62
mutant_R1.fastq mutant-no_snps.gff-5520/1 TCTCAGCACATAAATAATGTGGCAGCACAAT 63
mutant_R1.fastq mutant-no_snps.gff-5480/1 TCTTTTTTATCTTCGGTTAATTTATTATATT 0
mutant_R2.fastq mutant-no_snps.gff-4756/2 AATATAATAAATTAACCGAAGATAAAAAAGA 113
mutant_R1.fastq mutant-no_snps.gff-3476/1 ATCTCAGCACATAAATAATGTGGCAGCACAA 34
mutant_R1.fastq mutant-no_snps.gff-3476/1 TCTCAGCACATAAATAATGTGGCAGCACAAT 35
mutant_R1.fastq mutant-no_snps.gff-2218/1 TCTTTTTTATCTTCGGTTAATTTATTATATT 51
mutant_R2.fastq mutant-no_snps.gff-2152/2 ATCTCAGCACATAAATAATGTGGCAGCACAA 95
mutant_R2.fastq mutant-no_snps.gff-2152/2 TCTCAGCACATAAATAATGTGGCAGCACAAT 96
mutant_R1.fastq mutant-no_snps.gff-868/1 ATTGTGCTGCCACATTATTTATGTGCTGAGA 8
mutant_R1.fastq mutant-no_snps.gff-868/1 TTGTGCTGCCACATTATTTATGTGCTGAGAT 9
#
#Number of patterns found: 6/6 (100.00 %)
#Pattern Count
#AATATAATAAATTAACCGAAGATAAAAAAGA 4
#ATCTCAGCACATAAATAATGTGGCAGCACAA 7
#ATTGTGCTGCCACATTATTTATGTGCTGAGA 5
#TCTCAGCACATAAATAATGTGGCAGCACAAT 7
#TCTTTTTTATCTTCGGTTAATTTATTATATT 8
#TTGTGCTGCCACATTATTTATGTGCTGAGAT 5
#
#Total number of records searched: 24960
#Total number of characters searched: 3744000
#Total number of hits: 36
#Number of distinct records with a hit: 24
#
#Total number of hits in file 1: 13
#Total number of hits in file 2: 23
#Number of distinct records with a hit in file 1: 9
#Number of distinct records with a hit in file 2: 15
#Total number of extracted records: 48
Analyzing match statistics
Now we have the extracted sequencing read pairs (those were either mate contained any k-mers) in the files mutant_extracted_1.fastq and mutant_extracted_2.fastq in the output/ directory. Additionally, we have stored the same match statistics that just got printed to the screen in the file mutant_extracted.stats.json in the logs/ folder, but in JSON format instead of plain text. Because it is structured, the JSON file can easily be parsed by other tools or programming languages, making it simple to be used with e.g. Python or R.
A simple yet powerful command-line processor for JSON files is jq. We can use it to inspect the statistics in more detail, for instance to only print the summary statistics:
jq '.summary_statistics' logs/mutant_extracted.stats.json
{
"number_of_characters_searched": 3744000,
"number_of_distinct_records_with_a_hit": 24,
"number_of_matches": 36,
"number_of_patterns_found": 6,
"number_of_patterns_searched": 6,
"number_of_records_searched": 24960
}
If we wanted to do k-mer abundance analysis, we can rank k-mers by frequency:
jq -r '.pattern_hit_counts | to_entries | sort_by(.value) | reverse | .[] | "\(.value)\t\(.key)"' logs/mutant_extracted.stats.json
8 TCTTTTTTATCTTCGGTTAATTTATTATATT
7 TCTCAGCACATAAATAATGTGGCAGCACAAT
7 ATCTCAGCACATAAATAATGTGGCAGCACAA
5 TTGTGCTGCCACATTATTTATGTGCTGAGAT
5 ATTGTGCTGCCACATTATTTATGTGCTGAGA
4 AATATAATAAATTAACCGAAGATAAAAAAGA
Basic calculations are also possible, like calculating the percentage of sequencing reads containing any of our k-mers (note that when extracting read pairs, if at least one mate contains a query k-mer, the number of extracted reads can be higher than the number of matched reads):
jq -r '.summary_statistics | "Extraction rate: \((.number_of_distinct_records_with_a_hit/.number_of_records_searched)*100*1000|round/1000)/100% of reads contained target k-mers"' logs/mutant_extracted.stats.json
Extraction rate: 0.096/100% of reads contained target k-mers
Finally, a more advanced jq command that calculates the hit density per read file, i.e., the average number of matches in extracted sequences, rounded to two decimal places:
jq -r '.paired_end_reads_statistics |
"R1 density: \((.number_of_hits_in_file_1/.number_of_distinct_records_with_a_hit_in_file_1)*100|round/100) hits/read
R2 density: \((.number_of_hits_in_file_2/.number_of_distinct_records_with_a_hit_in_file_2)*100|round/100) hits/read"' \
logs/mutant_extracted.stats.json
R1 density: 1.44 hits/read
R2 density: 1.53 hits/read
Aligning the extracted reads
Now we can map the extracted reads back to the wildtype reference genome, for instance using bowtie2. As a prerequisite, we have to generate an index for our reference:
bowtie2-build data/wildtype.fna data/wildtype
The extracted reads are aligned with the --very-sensitive-local parameter to allow soft-clipping the reads for alignment:
bowtie2 -x data/wildtype -1 output/mutant_extracted_1.fastq -2 output/mutant_extracted_2.fastq --very-sensitive-local > output/mutant_extracted.sam
Tagging the aligned reads with k-mers
It is often a requirement to sort the SAM file by coordinates before doing follow-up analysis. This can be done using samtools:
samtools sort output/mutant_extracted.sam -O sam > output/mutant_extracted.sorted.sam
We would like to know which k-mers the aligned reads contain. To do so, we can use MerKurio’s tag subcommand. Again, we are using the -r flag to also search for reverse complements:
merkurio tag -i output/mutant_extracted.sorted.sam -o output/mutant_extracted.sorted.tagged.sam -f data/significant_kmers.txt -r
Inspecting the SAM file shows us that the reads are mapped to a region from around 45,000 to 49,000. The k-mers contained in aligned sequences are stored under the km tag (Z denotes a string type, in accordance with the official SAM field specifications):
less output/mutant_extracted.sorted.tagged.sam
Note that we could have also mapped all reads to the reference genome and filter the resulting SAM file to keep only those sequences containing k-mers with merkurio tag -m [...], but in a real-life example with up to terabytes of raw sequencing reads this can become unfeasible, and we would not keep the mates of reads with our k-mers. Extracting only the relevant reads before mapping can save time and disk space.
Visualization of tagged reads
The aligned and tagged sequencing reads can be visualized in the Integrative Genome Browser (IGV). Start IGV (by typing igv in the terminal) and load the wildtype.fna genome, the file with the gene annotations wildtype.gff, and the tagged reads mutant_extracted.sorted.tagged.sam.
To do so, in the menu select “Genomes → Load Genome from File…” and “File → Load from File…”, respectively. IGV might prompt that it must first generate an index for some files.
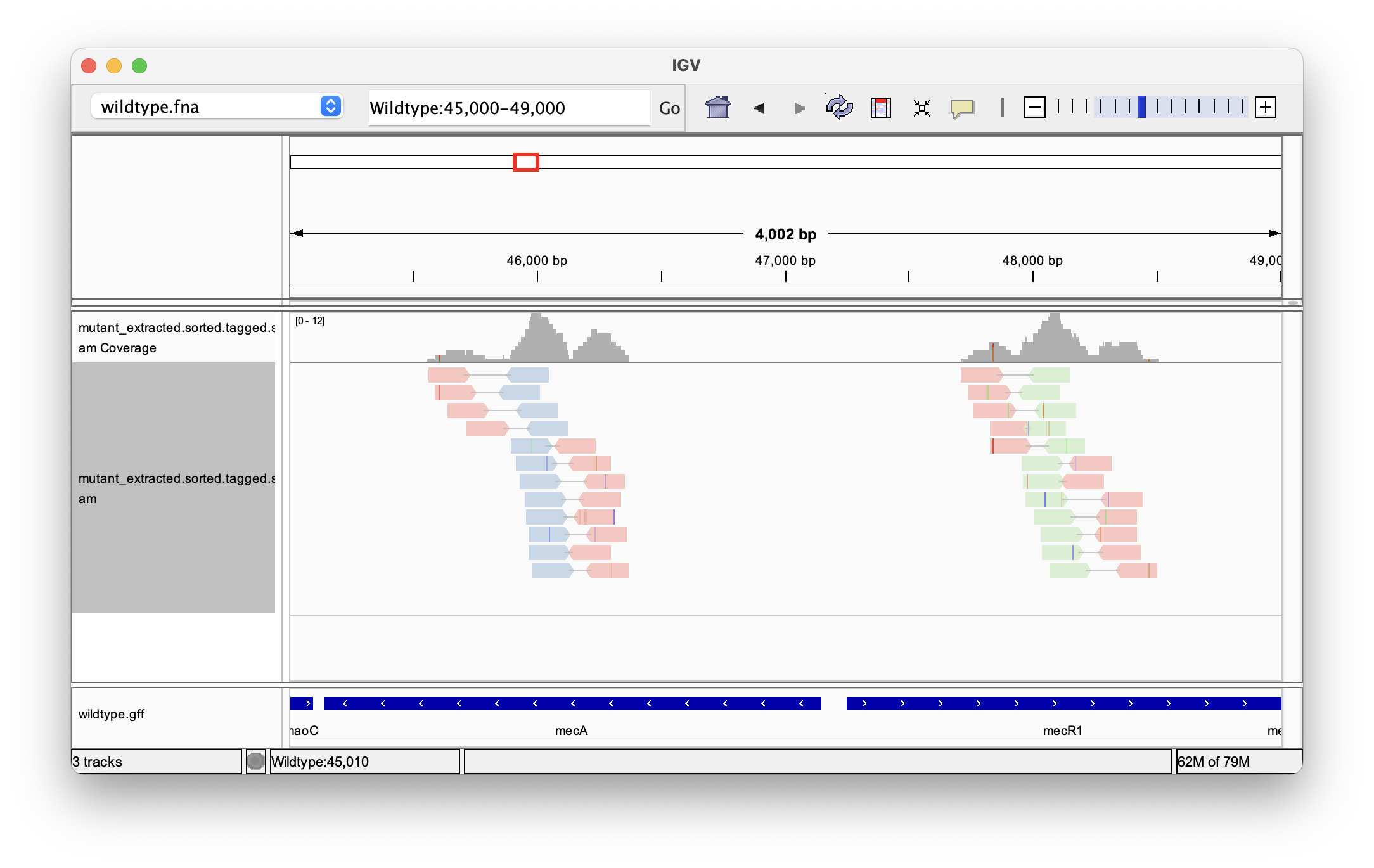
Now, zoom in on the region of interest by entering the coordinates and clicking “Go”: Wildtype:45,000-49,000. Right-clicking on the track with the aligned reads and selecting “View as pairs” to group the read pairs. An advantage of our tagged reads is that we can highlight the presence of specific k-mers. To do so, press right click on the track, go “Color alignments by → tag” and enter km:
Clicking on a single read also shows us its k-mer tags. When looking for specific k-mers, it can also make sense to group them by tag:
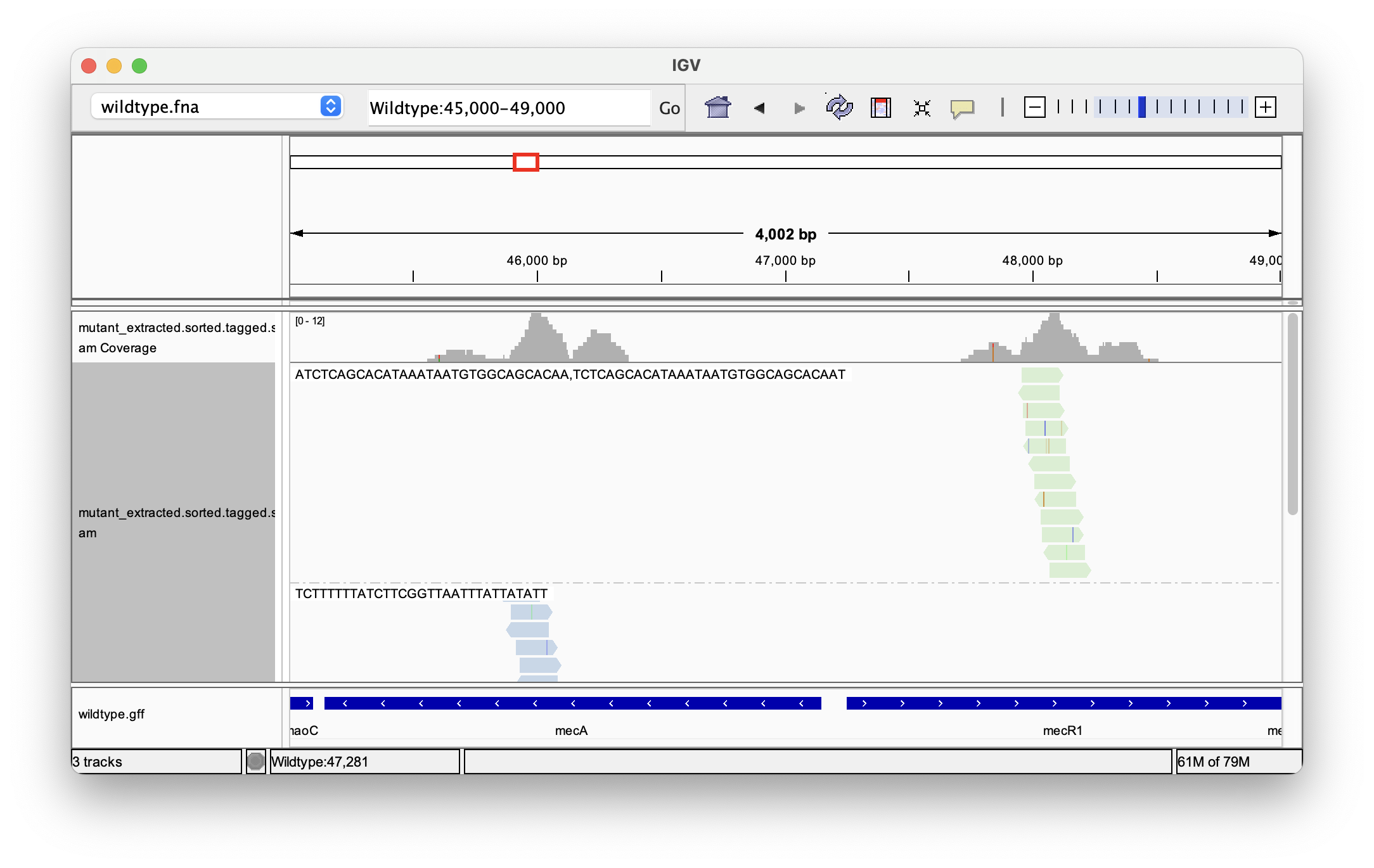
By looking at the track with the gene annotations on the bottom, we can see that the reads mapped to positions in the genes mecA and mecR1, which are expressing a penicillin-binding protein and a methicillin-resistance regulatory protein, respectively. By extracting sequencing reads significantly associated with antibiotics resistance and mapping them back to the reference genome we were able to successfully identify candidate genes for this trait.
Follow-up analysis could include looking for variants (SNPs and indels) present in the tagged reads, or intersecting the gene annotation with the positions of extracted reads.
Feature Overview
MerKurio provides two complementary subcommands:
- 🔍 Extract: Search FASTA/FASTQ data for k-mers and write records with matching k-mers to the terminal or a new file.
- Supports paired-end reads (a hit in one read extracts the whole pair).
- 📑 Tag: Annotate BAM/SAM alignments with k-mer tags and filter them based on matching k-mers.
- Adds a two‑letter tag (default
km) with comma-separated matching k‑mers (follows the SAM format specification). - Optionally keeps only reads containing at least one k‑mer.
- Multithreaded processing when working with BAM files.
- Adds a two‑letter tag (default
Both commands share additional features:
- Records detailed matching statistics (positions of k-mer occurence, summary statistics, metadata).
- Human readable output in plain text.
- Structured JSON logs for easy machine parsing.
- Reads compressed input files (
.gz,.bz2,.xz). - Can seach for reverse complements or only canonical forms of k-mers.
- Case-insensitive search or conversion to lower-/uppercase.
- Inverse matching to keep only those records without matches.
- Query k-mers can be provided as command line arguments or in a file (FASTA or plain text).
- File types are inferred automatically.
- Record output can be suppressed to only record statistics.
The extract Subcommand
Running merkurio extract will extract records from a FASTA or FASTQ sequence file based on a list of query sequences (k-mers). The extracted records are written to the terminal or to a new file in the same format as the input file. The k-mers can be provided as a list of strings on the command line or in a file (FASTA or plain text file).
Detailed match statistics are written to the terminal (stdout) or to a file if a path is provided, showing which records got matched by which patterns, including the zero-based start of the position of the match. Matching statistics can also be saved in JSON format for parsing by other programs.
MerKurio supports input files compressed with gzip, bzip2 or xz. Note that searching with MerKurio is case-sensitive by default, but can be set to ignore the case. Alternatively, all input k-mers can be converted to lower- or uppercase.
MerKurio supports processing of paired-end reads, where a hit in one read also extracts the other read of that pair. The extracted records are written to separate files, with their names being set accordingly. In this mode, additional statistics are provided.
Further options include the ability to also search for the reverse complements of nucleotide query sequences (i.e., a sequence is reversed, and C/G and A/T are swapped), or searching for canonical k-mers only. Output of matching records can be suppressed if only the statistics are needed. MerKurio tries to select the most efficient algorithm for the given query sequences, but the user can override this choice by selecting a specific algorithm (not recommended).
For a detailed explanation of the matching statistics, see the Log Output section.
Usage
Basic usage of the extract subcommand is as follows:
merkurio extract [OPTIONS] -i <IN_FASTX> <-s <KMER_SEQ>...|-f <KMER_FILE>>
For more examples, see the Example Usage section.
Detailed Description of the Available Options
You can display a help message with -h or --help.
Input/output parameters:
| Short flag | Long flag | Description |
|---|---|---|
-i, -1 | --in-fastx | <Path to input file (FASTA/FASTQ)> Supports .gz, .bz2, and .xz compressed files. |
-s | --kmer-seq | <Query sequences to search for>... Multiple sequences can be provided as arguments, separated by spaces. Duplicate sequences are ignored. |
-f | --kmer-file | <Path to a file containing query sequences> Can be in FASTA format or plain text, with empty lines and lines preceded by a # being ignored. |
-o | --out-fastx | <Output file path> If not provided, output is written to stdout (i.e., the terminal). The correct file extension is added automatically. |
-2 | --in-fastq-2 | <Path to the second input file for paired-end reads> When processing paired-end reads, a match in one read of the pair will extract both. Output is written to two separate files, appending _1 and _2 to the base names of the output files. |
-l | --out-log | Set this flag without any arguments to write matching statistics to stdout, or write to file if a path to the output file is passed as an argument to this option. For an explanation of the matching statistics, see the section below. |
-j | --json-log | Set this flag without any arguments to write matching statistics in JSON format to stdout, or provide a file path to write JSON log to a file. If both -l and -j are set without arguments, it will return an error. |
-S | --suppress-output | Set this flag to suppress the output of matching records. Only the matching statistics are printed (either use -l or -j for plain text or JSON logging, respectively). |
Search parameters:
| Short flag | Long flag | Description |
|---|---|---|
-r | --reverse-complement | Set this flag to also search for the reverse complements of the nucleotide query sequences (i.e., a sequence is reversed, and C/G and A/T are swapped). Duplicate sequences are not added to the list. Works for IUPAC codes, everything else is unchanged. |
-c | --canonical | Set this flag to only search for the canonical forms of k-mers (i.e., the lexicographically first between a sequence and its reverse complement). |
-v | --invert-match | Set this flag to invert the matching behavior: instead of keeping records that contain matching k-mers, keep only records that do not match any of the k-mers. |
-I | --case-insensitive | Set this flag to use case-insensitive matching. Always uses the Aho-Corasick algorithm. |
-L | --lowercase | Set this flag to convert all input sequences to lowercase. |
-U | --uppercase | Set this flag to convert all input sequences to uppercase. |
Special parameters:
| Short flag | Long flag | Description |
|---|---|---|
-a | --aho-corasick | Set this flag to use the Aho-Corasick algorithm. It is best used when searching for lots of patterns at once or when searching for k-mers with more than 64 characters, which are too long for the BNDMq algorithm. |
-q | --q-size | <Size of q-grams for BNDMq> Use the Backwards Nondeterministic DAWG Matching algorithm tuned with q-grams (BNDMq) with this value for the size of q. The optimal value of q depends on the size of the query and type of text searched and is usually around 3-5. It must not be larger than the length of the pattern. |
Example Usage
An example usage of the extract subcommand to extract records from a FASTA file based on a list of k-mers and their reverse complements is shown below. Records from input.fasta matching any of the k-mers in query_kmers.txt are written to the file output.fasta. Logging information is written to the terminal (stdout):
merkurio extract -i input.fasta -o output.fasta -f query_kmers.txt -r -l
Another example where paired-end reads are extracted if they contain the sequence ACGT or TGCA; the extracted records are printed to the terminal and logging information is written to two files; as plain text to log.txt, and in JSON format to log.json:
merkurio extract -1 in_R1.fastq -2 in_R2.fastq -o output -s ACGT TGCA -l log.txt -j log.json
The following example only writes the matching statistics to a JSON file log.json; FASTA output is suppressed:
merkurio extract -i input.fasta -f query_kmers.txt -S -j log.json
The tag Subcommand
Running merkurio tag will tag aligned sequences in a SAM/BAM file with contained k-mers. If a record contains one or more of the k-mers, it is annotated with a tag (“km” by default; must be exactly two characters long) and the respective k-mers, separated by commas.
The k-mers can be provided as a list of strings on the command line or in a file (FASTA or plain text file). The output is written to a SAM or BAM file, depending on the file extension. If the chosen tag is already present in the records, the new values are added to existing ones. Optionally, keep only records which are matching at least one k-mer, or keep only records which are not matching any k-mer. Multithreading is supported for parsing BAM files.
Note that searching with MerKurio is case-sensitive by default, but can be set to ignore the case. Alternatively, all input k-mers can be converted to lower- or uppercase.
Detailed match statistics are written to the terminal (stdout) or to a file if a path is provided, showing which records got matched by which patterns, including the zero-based start of the position of the match. Matching statistics can also be saved in JSON format for parsing by other programs.
Further options include the ability to also search for the reverse complements of nucleotide query sequences (i.e., a sequence is reversed, and C/G and A/T are swapped), or searching for canonical k-mers only. Output of matching records can be suppressed if only the statistics are needed. MerKurio tries to select the most efficient algorithm for the given query sequences, but the user can override this choice by selecting a specific algorithm (not recommended).
For a documentation of the SAM format, see the SAM format specification.
For a documentation on the SAM/BAM tags, see the SAM optional fields specification. The default tag is km:Z:, followed by the k-mers separated by commas. This meets the specification for a string tag and lowercase characters are reserved for user-defined tags.
For a detailed explanation of the matching statistics, see the Log Output section.
Usage
Basic usage of the tag subcommand is as follows:
merkurio tag [OPTIONS] -i <IN_FILE> <-s <KMER_SEQ>...|-f <KMER_FILE>>
For more examples, see the Example Usage section.
Detailed Description of the Available Options
You can display a help message with -h or --help.
Input/output parameters:
| Short flag | Long flag | Description |
|---|---|---|
-i | --in-file | <Path to input file (SAM/BAM)> Supports .bam and .sam files, automatically recognizing the file type based on the extension. |
-s | --kmer-seq | <Query sequences to search for>... Multiple sequences can be provided as arguments, separated by spaces. Duplicate sequences are ignored. |
-f | --kmer-file | <Path to a file containing query sequences> Can be in FASTA format or plain text, with empty lines and lines preceded by a # being ignored. |
-o | --out-file | <Output file path> If not provided, output is written to stdout (i.e., the terminal). The extension of the output file path determines the file type (SAM/BAM). If none is provided, the input file type will be used. |
-l | --out-log | Set this flag without any arguments to write matching statistics to stdout, or write to file if a path to the output file is passed as an argument to this option. For an explanation of the matching statistics, see the section below. |
-j | --json-log | Set this flag without any arguments to write matching statistics in JSON format to stdout, or provide a file path to write JSON log to a file. If both -l and -j are set without arguments, it will return an error. |
-S | --suppress-output | Set this flag to suppress the output of matching records. Only the matching statistics are printed (either use -l or -j for plain text or JSON logging, respectively). |
-m | --filter-matching | Set this flag to only output records that contain at least one of the k-mers. If no k-mers are found, the record is not output. |
Search parameters:
| Short flag | Long flag | Description |
|---|---|---|
-r | --reverse-complement | Set this flag to also search for the reverse complements of the nucleotide query sequences (i.e., a sequence is reversed, and C/G and A/T are swapped). Duplicate sequences are not added to the list. Works for IUPAC codes, everything else is unchanged. |
-c | --canonical | Set this flag to only search for the canonical forms of k-mers (i.e., the lexicographically first between a sequence and its reverse complement). |
-v | --invert-match | Set this flag to invert the matching behavior: instead of keeping records that contain matching k-mers, keep only records that do not match any of the k-mers. This flag cannot be used together with -m (–filter-matching). |
-I | --case-insensitive | Set this flag to use case-insensitive matching. Always uses the Aho-Corasick algorithm. |
-L | --lowercase | Set this flag to convert all input sequences to lowercase. |
-U | --uppercase | Set this flag to convert all input sequences to uppercase. |
Special parameters:
| Short flag | Long flag | Description |
|---|---|---|
-t | --tag | <Tag to use> The tag must be exactly two characters long. The default is km. Consider the SAM specifications when choosing a tag to avoid conflicts. If a tag is already present, the new values are appended to existing ones. |
-p | --threads | <Number of threads> The number of parallel threads used when processing BAM files. Default is 1. Has no effect on SAM files. |
-a | --aho-corasick | Set this flag to use the Aho-Corasick algorithm. It is best used when searching for lots of patterns at once or when searching for k-mers with more than 64 characters, which are too long for the BNDMq algorithm. |
-q | --q-size | <Size of q-grams for BNDMq> Use the Backwards Nondeterministic DAWG Matching algorithm tuned with q-grams (BNDMq) with this value for the size of q. The optimal value of q depends on the size of the query and type of text searched and is usually around 3-5. It must not be larger than the length of the pattern. |
Example Usage
An example usage of the tag subcommand to tag a BAM file with the k-mers in the file query_kmers.fasta and output a SAM file is shown below:
merkurio tag -i input.bam -o output.sam -f query_kmers.fasta
Another example where the k-mers are provided on the command line and the search is also performed for their reverse complements. The tag is set to “MK”. BAM file processing is done with 4 threads:
merkurio tag -i input.bam -o output.bam -s ACGT TGCA -r -p 4 -t MK
This example shows how to filter the output to only include records that contain at least one of the k-mers. Output is written to the terminal (stdout):
merkurio tag -i input.bam -s AAATCAAGA -m
The next example only writes the matching statistics to a JSON file log.json; SAM output is suppressed:
merkurio tag -i input.bam -s AAATCAAGA -S -j log.json
Explanation of the Log Matching Statistics
Recording detailed match statistics is available for both the extract and tag subcommands. The logs of the two subcommands are mostly identical, with the exception of the paired_end_reads_statistics section, which is only present in the extract subcommand log after processing paired-end reads, and an additional field tag in the JSON log of the tag subcommand. The examples on this page are based on the extract subcommand.
There are two types of log output format available: plain text and JSON. An example of each is provided below. Except for minor differences in formatting, the information contained in both log formats is the same. Plain text and JSON logs are written to files, provided with the -l and -j arguments, respectively. If only the flag are present, the log will be written to the terminal (stdout).
When processing paired-end reads, additional statistics are collected. If a single file is processed, the number of extracted records equals the number of distinct records with a hit. In paired-end reads, however, a match in one read of a pair will extract both of them.
Plain Text Log
The log in plain text format is meant to be human-readable. Lines starting with a # contain meta-information and matching statistics. The log also contains a tab-delimited table with an entry for each pattern found in a record, listing the file name, record ID, query sequence and zero-based position of the match. These lines are not prefixed with a # to facilitate inspection and further analysis, e.g. using grep -v '^#' to extract a tab-delimited table.
After the table, patterns and their number of occurences are listed. This is followed by summary statistics.
#MerKurio extract log
#2024-09-23T00:13:17+02:00[Europe/Vienna]
#Running MerKurio version 1.0.0
#Command line: merkurio extract -i reads_1.fastq -2 reads_2.fastq -s ATCG -l -r
#Searching for 2 patterns
#
#File Record Pattern Position (zero-based)
reads_1.fastq record 1/1 ATCG 0
reads_2.fastq record 1/2 CGAT 147
reads_1.fastq record 57/1 ATCG 13
#
#Number of patterns found: 2/2 (100.00 %)
#Pattern Count
#ATCG 2
#CGAT 1
#
#Total number of records searched: 10000
#Total number of characters searched: 150000
#Total number of hits: 3
#Number of distinct records with a hit: 3
When processing paired-end reads, the log will contain an additional block of statistics:
#
#Total number of hits in file 1: 2
#Total number of hits in file 2: 1
#Number of distinct records with a hit in file 1: 2
#Number of distinct records with a hit in file 2: 1
#Total number of extracted records: 4
JSON Log
The log in JSON format is intended to be machine-readable. It contains the same information as the plain text log but in a structured format. The JSON log contains five main sections: matching_records, summary_statistics, meta_information, paired_end_reads_statistics, and pattern_hit_counts.
The matching_records array contains a list of all matches. Each match is stored as an object with the file name, record ID, query sequence, and zero-based position of the match in that record.
The summary_statistics object contains the total number of records searched, the total number of characters of sequences searched, the total number of hits, the number of records with at least one hit, and the numbers of searched/matching patterns.
The meta_information object contains the command passed to execute MerKurio as an array, the program’s name (MerKurio) and version, the timestamp when the log was generated, and the SAM tag in case of the tag subcommand. It also stores the names of input files in an object and information about the search mode (inverted matching extracts only non-matching records, case-insensitive search, used algorithm).
The paired_end_reads_statistics object contains information about the number of hits in each file, the number of records with at least one hit for each file, and the total number of extracted records. In paired-end read mode, a match in one read of a pair will extract both of them. The total number of extracted records can thus be higher than the number of distinct records with a hit. The searching_paired_end_reads boolean indicates whether paired-end reads were used (i. e., a second read file was passed via the -2 flag).
The pattern_hit_counts field is a dictionary, with an entry for each pattern searched and the number of times it was found.
{
"matching_records": [
{
"file": "reads_1.fastq",
"pattern": "ATCG",
"position": "0",
"record_id": "record 1/1"
},
{
"file": "reads_2.fastq",
"pattern": "CGAT",
"position": "147",
"record_id": "record 1/2"
},
{
"file": "reads_1.fastq",
"pattern": "ATCG",
"position": "13",
"record_id": "record 1/1"
}
],
"meta_information": {
"case_insensitive": false,
"command_line": [
"merkurio",
"extract",
"-i",
"reads_1.fastq",
"-2",
"reads_2.fastq",
"-s",
"ATCG",
"-r",
"-j",
"log.json"
],
"input_files": {
"kmer_file": null,
"record_file_1": "reads_1.fastq",
"record_file_2": "reads_2.fastq"
},
"inverted_matching": false,
"program": "merkurio",
"search_algorithm": "BNDMq",
"subcommand": "extract",
"timestamp": "2024-09-23T00:13:17+02:00[Europe/Vienna]",
"version": "1.0.0"
},
"paired_end_reads_statistics": {
"number_of_distinct_records_with_a_hit_in_file_1": 2,
"number_of_distinct_records_with_a_hit_in_file_2": 1,
"number_of_extracted_records": 4,
"number_of_hits_in_file_1": 2,
"number_of_hits_in_file_2": 1,
"searching_paired_end_reads": true
},
"pattern_hit_counts": {
"ATCG": 2,
"CGAT": 1
},
"summary_statistics": {
"number_of_characters_searched": 150000,
"number_of_distinct_records_with_a_hit": 3,
"number_of_matches": 3,
"number_of_patterns_found": 2,
"number_of_patterns_searched": 2,
"number_of_records_searched": 10000
}
}
This JSON output can easily be read by other programming languages like Python, or processed with other command-line tools such as jq. For tutorials on both, see the next sections.
Python Analysis of the JSON Log
Python is a general purpose programming language with an easy-to-learn syntax.
It natively supports parsing JSON files with the json module. Replace log.json with the path of your log file. It will store the JSON log as a standard Python dictionary:
import json
# Load the JSON log
with open('log.json') as f:
data = json.load(f)
# Accessing the "summary_statistics" entry
data["summary_statistics"]
Example: Sorting Records by Match Count
In this example, the JSON log is parsed and the match counts per record are stored in a dictionary. Then, the entries are printed in descending order:
import json
from collections import defaultdict
# Load the JSON log
with open("log.json") as f:
data = json.load(f)
# Count matches per distinct record
record_counts = defaultdict(int)
for entry in data["matching_records"]:
record_id = entry["record_id"]
record_counts[record_id] += 1
# Sort by match count, descending
sorted_records = sorted(record_counts.items(), key=lambda x: x[1], reverse=True)
# Print results
for record_id, count in sorted_records:
print(f"{record_id}: {count} match{'es' if count > 1 else ''}")
Example: Plot a Histogram of Match Densities
Using the external dependency Matplotlib, a histogram of match position density can be plotted:
import json
import matplotlib.pyplot as plt
# Load the JSON log
with open('log.json') as f:
data = json.load(f)
# Extract and convert positions to integers
positions = [int(entry["position"]) for entry in data["matching_records"]]
# Plot histogram
plt.hist(positions)
plt.xlabel("Pattern match position in record")
plt.ylabel("Frequency")
plt.title("Histogram of Pattern Match Positions")
plt.tight_layout()
plt.show()
Useful jq Commands
jq is a lightweight and flexible command-line JSON processing language.
A work-in-progress list of jq commands to analyse the JSON logs of MerKurio on the command line. Replace log.json with the path to your JSON file.
Quality metrics
For paired-end reads:
jq -r '. | "Number of reads with matches: \(.matching_statistics.number_of_distinct_records_with_a_hit)
Number of extracted reads: \(.paired_end_reads_statistics.number_of_extracted_records)
Total number of reads: \(.matching_statistics.number_of_records_searched)
Match rate: \((.matching_statistics.number_of_distinct_records_with_a_hit/.matching_statistics.number_of_records_searched)*100*1000|round/1000)/100% of reads contained target k-mers
Extraction rate: \((.paired_end_reads_statistics.number_of_extracted_records/.matching_statistics.number_of_records_searched)*100*1000|round/1000)/100% of reads were extracted"' log.json
Summary reports
For single files:
jq -r '
"--- MerKurio Extraction Summary ---",
"Run: " + .meta_information.timestamp,
"Algorithm: " + .meta_information."search-algorithm",
"",
"SEARCH RESULTS:",
"• Patterns searched: " + (.matching_statistics.number_of_patterns_searched | tostring),
"• Patterns found: " + (.matching_statistics.number_of_patterns_found | tostring) + " (" + ((.matching_statistics.number_of_patterns_found/.matching_statistics.number_of_patterns_searched)*100*100 | round/100 | tostring) + "%)",
"• Total matches: " + (.matching_statistics.number_of_matches | tostring),
"• Reads with hits: " + (.matching_statistics.number_of_distinct_records_with_a_hit | tostring) + "/" + (.matching_statistics.number_of_records_searched | tostring),
""
' log.json
For paired-end reads:
jq -r '
"--- MerKurio Extraction Summary ---",
"Run: " + .meta_information.timestamp,
"Algorithm: " + .meta_information."search-algorithm",
"",
"SEARCH RESULTS:",
"• Patterns searched: " + (.matching_statistics.number_of_patterns_searched | tostring),
"• Patterns found: " + (.matching_statistics.number_of_patterns_found | tostring) + " (" + ((.matching_statistics.number_of_patterns_found/.matching_statistics.number_of_patterns_searched)*100*100 | round/100 | tostring) + "%)",
"• Total matches: " + (.matching_statistics.number_of_matches | tostring),
"• Reads with hits: " + (.matching_statistics.number_of_distinct_records_with_a_hit | tostring) + "/" + (.matching_statistics.number_of_records_searched | tostring),
"",
"PAIRED-END DETAILS:",
"• R1 hits: " + (.paired_end_reads_statistics.number_of_hits_in_file_1 | tostring) + " in " + (.paired_end_reads_statistics.number_of_distinct_records_with_a_hit_in_file_1 | tostring) + " reads",
"• R2 hits: " + (.paired_end_reads_statistics.number_of_hits_in_file_2 | tostring) + " in " + (.paired_end_reads_statistics.number_of_distinct_records_with_a_hit_in_file_2 | tostring) + " reads",
"• Extracted pairs: " + (.paired_end_reads_statistics.number_of_extracted_records | tostring),
""
' log.json
Data export
K-mers with at least one match to a FASTA file, with the match count in the sequence header:
jq -r '
.pattern_hit_counts
| to_entries
| map(select(.value > 0))
| to_entries
| map(">kmer"+((.key + 1)|tostring)+"|count="+(.value.value|tostring)+"\n"+.value.key)
| .[]
' log.json > matched_kmers.fasta
Pattern hit counts to TSV:
jq -r '.pattern_hit_counts | to_entries | ["Pattern", "Count"], (.[] | [.key, .value]) | @tsv' log.json > kmer_counts.tsv
Pattern positions to TSV:
jq -r '.matching_records | ["Pattern", "Position", "File", "ReadID"], (.[] | [.pattern, .position, .file, .record_id]) | @tsv' log.json > kmer_positions.tsv
Multiple hits of the same patterns
Producing compact output (empty if nothing is found):
jq -r '
.matching_records
| group_by(.pattern)
| .[]
| . as $pattern_group
| ($pattern_group | group_by(.record_id) | map(select(length > 1))) as $multi_hits
| if ($multi_hits | length) > 0 then
" Pattern: " + $pattern_group[0].pattern,
" Reads with multiple hits:",
($multi_hits[] | " • " + .[0].record_id + " (" + (length | tostring) + " hits at positions: " + ([.[].position | tonumber] | sort | map(tostring) | join(", ")) + ")")
else empty end
' log.json
Display information line-wise:
jq -r '
.matching_records
| group_by(.pattern)
| .[]
| . as $pattern_group
| ($pattern_group | group_by(.record_id) | map(select(length > 1))) as $multi_hits
| if ($multi_hits | length) > 0 then
"",
"=" * 60,
"PATTERN: " + $pattern_group[0].pattern,
"=" * 60,
($multi_hits[] |
"Read: " + .[0].record_id + " (File: " + .[0].file + ")",
"Hits: " + (length | tostring),
(. | sort_by(.position | tonumber) | .[] | " Position " + .position),
""
)
else empty end
' log.json
Less detailed output:
jq -r '
.matching_records
| group_by(.pattern)
| map({
pattern: .[0].pattern,
multi_hit_reads: (. | group_by(.record_id) | map(select(length > 1)) | length),
total_multi_hits: (. | group_by(.record_id) | map(select(length > 1)) | map(length) | add // 0)
})
| map(select(.multi_hit_reads > 0))
| if length > 0 then
.[] | .pattern[0:30] + "... : " + (.multi_hit_reads | tostring) + " read(s) with " + (.total_multi_hits | tostring) + " total multi-hits"
else
"No reads found with multiple hits of the same pattern"
end
' log.json